Introduction
The original motivation for this work was a WCS based replacement of Multidrizzle, now released as Astrodrizzle, and specifically a requirement for the availability and management of multiple WCS sets, complete with distortion, within one science file. However, the concept of encapsulating astrometric solutions is more general than that since each solution may represent a different astrometric alignment, either with a catalog or another image. Furthermore, computing accurate astrometric solutions requires considerable effort and time so having a way to distribute them, apply them to a science observation and switch between different WCSs efficiently would facilitate many aspects of data analysis.
Some of the immediate areas for the use of headerlets with HST data include the HST archive, the HLA and other legacy projects which provide improved astrometry of HST observations. The HST Archive, for example, provides access to all HST data, most of which can not be readily combined together due to errors in guide star astrometry imposing offsets between images taken using different pairs of guide stars. A lot of effort has gone into computing those offsets so that all the images taken at a particular pointing can be combined successfully with astrometry matched to external astrometric catalogs. Unfortunately, there is no current mechanism for passing those updated solutions along to the community without providing entirely new copies of all the data.
Source Image
Any science observation with a valid WCS described by the FITS standard or any of the WCS conventions implemented in pywcs may serve as a source for creating a headerlet.
We describe as an example the type of WCS information of a typical HST ACS/WFC image as it is distributed by the HST archive (OTFR) after being processed with the latest image calibrations, including applying the latest available distortion models. The full description of the WCS is available in the “Distortion Correction In HST Files” report [Hack] by Hack et al. The science header now contains the following set of keywords and extensions to fully describe the WCS with distortion:
Linear WCS keywords: CRPIX, CRVAL, CTYPE, CD matrix keywords
SIP coefficients: A_*_* and B_*_*, A_ORDER, B_ORDER
The first order coefficients from the IDC table: (needed by astrodrizzle) OCX10, OCX11, OCY10, and OCY11 keywords
- NPOL distortion: if an NPOLFILE has been specified for the image,
CPDIS and DP record-value keywords to point to WCSDVARR extensions (Distortion Paper [Calabretta] )
- Detector defect correction: (for ACS/WFC this is a column defect)if a D2IMFILE has been
specified for use with the image, the D2IMEXT, D2IMERR and AXISCORR keywords point to the D2IMARR extension
Some of the distortion information may be stored in additional extensions in the science file:
- WCSDVARR extensions: 2 extensions for each chip with lookup tables containing
the non-polynomial corrections, with each extension corresponding to an axis of the image (X correction or Y correction)
D2IMARR extension: an extension with a lookup table containing the row- or column-correction from the D2IMFILE.
Each science header will have its own set of these keywords and extensions that will be kept together as part of the headerlet definition. This avoids any ambiguity as to what solution was used for any given WCS.
An HST ACS/WFC exposure would end up with the following set of extensions:
EXT# FITSNAME FILENAME EXTVE DIMENS BITPI OBJECT
0 j8hw27c4q_flt j8hw27c4q_flt.fits 16
1 IMAGE SCI 1 4096x2048 -32
2 IMAGE ERR 1 4096x2048 -32
3 IMAGE DQ 1 4096x2048 16
4 IMAGE SCI 2 4096x2048 -32
5 IMAGE ERR 2 4096x2048 -32
6 IMAGE DQ 2 4096x2048 16
7 IMAGE D2IMARR 1 4096 -32
8 IMAGE WCSDVARR 1 65x33 -32
9 IMAGE WCSDVARR 2 65x33 -32
10 IMAGE WCSDVARR 3 65x33 -32
11 IMAGE WCSDVARR 4 65x33 -32
These additional extensions add approximately 100kB to a typical ACS/WFC image making them a space efficient means of managing all the distortion and WCS information.
Headerlet Definition
A headerlet is a self-consistent definition of a single WCS
including all distortion for all chips/detectors of a single exposure.
This is different from alternate WCS defined in Greisen, E. W., and Calabretta (Paper I) [Greisen]
in that by definition all alternate WCSs share the same distortion model while headerlets
may be based on different distortion models. A headerlet does not include alternate WCSs.
It is stored as a multi-extension FITS file following the structure of the science file.
The WCS information in the science header is saved in the header of an HDU with EXTNAME ‘SIPWCS’.
All other HDUs in the headerlet (containing distorion information)
have the same EXTNAME as the science file.
SIPWCS - A New FITS Extension
We introduce a new HDU with EXTNAME SIPWCS. It has no data and the header
contains all the WCS-related keywords from the SCI
header. As a minimum it contains the basic WCS keywords described in Paper I [Greisen]
If the science observation has a SIP distortion model, the SIP keywords are included
in this extension. If the distortion includes a non-polynomial
part, the keywords describing the extensions with the lookup tables
(EXTNAME=WCSDVARR) are also in this header. If there’s a detector defect correction
(row or column correction), the keywords describing the D2IMARR HDU are also in this
header. In addition, each SIPWCS header contains two keywords which point back to the HDU
of the original science file which was the source for it. These keywords are TG_ENAME and
TG_EVER and have the meaning of (extname, extver) for the data extension in the science file.
The keywords in this extension are used by the software to overwrite the keywords in the corresponding SCI header to update the WCS solution for each chip without any computation.
Headerlet File Structure
This SIPWCS extension along with all WCSDVARR extensions and the D2IMARR extension if available
fully describe the WCS of each chip.
The listing of the FITS extensions for a headerlet for a sample ACS/WFC exposure after writing
it out to a file is:
EXT# FITSNAME FILENAME EXTVE DIMENS BITPI OBJECT
0 j8hw27c4q j8hw27c4q_hdr.fits 16
1 IMAGE SIPWCS 1 8
2 IMAGE SIPWCS 2 8
3 IMAGE WCSDVARR 1 65x33 -32
4 IMAGE WCSDVARR 2 65x33 -32
5 IMAGE WCSDVARR 3 65x33 -32
6 IMAGE WCSDVARR 4 65x33 -32
7 IMAGE D2IMARR 1 4096 -32
Note
A headerlet derived from a full-frame WFC3/UVIS image would only contain a PRIMARY header and two SIPWCS extensions (one for each SCI extension) as WFC3/UVIS does not currently have non-polynomial distortion or any detector defect corrections.
The keywords used to populate the headerlet come from all the extensions of the updated FITS file, as illustrated in the following figure.
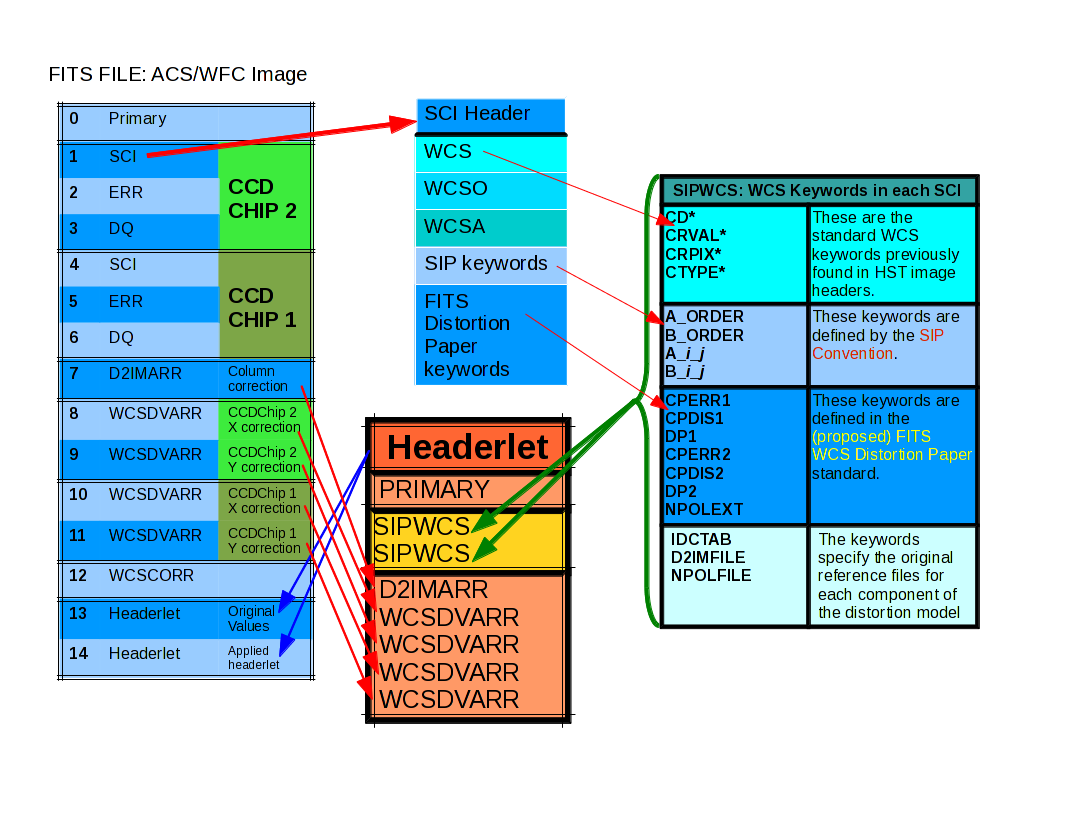
This figure shows the keywords that are included in a headerlet, the extensions included in a headerlet, and how a headerlet appears as a new extension when it gets appended to the original ACS/WFC file.
Headerlet Primary Header
The list below contains all keywords specific to the primary header of a headerlet with a brief description how to determine their value. Note that all keywords will be present in the header and ‘required’ and ‘optional’ below refers to their value.
HDRNAME- (required) a unique name for the headerlet
the value is given by the user as a parameter to
create_headerletorwrite_headerletHDRNAME<wcskey> from the science file is used
WCSNAME<wcskey> from the science file is used
KeyError is raised
DESTIM- (required) target image filename - used to determine if a headerlet can be applied to a science file.
the ROOTNAME keyword of the original science file
the name of the science file
WCSNAME- (required) name for the WCS
the value is given by the user as a parameter to
create_headerletorwrite_headerletWCSNAME<wcskey> from the science file is used
the value of hdrname parameter is used
HDRNAME<wcskey> from the science file
KeyError is raised
DISTNAME- (optional) name of distortion model
- The value of DISTNAME has the form
<idctab rootname>-<npolfile rootname>-<d2imfile rootname> and has a value of ‘NONE’ if no reference files are specified.
SIPNAME- (optional) name of SIP model
SIPNAME is constructed as <ROOTNAME>_<IDCTAB_rootname>, where
ROOTNAME is the keyword from the science file header (or the file name)
IDCTAB_rootname is the rootname of the idctab file
so for example, SIPNAME for a science file j94f05bgq_flt.fits and an idctab file postsm4_idc.fits is j94f05bgq_postsm4
If the SIP coefficients are present in the header but IDCTAB is missing or invalid, then SIPNAME is set to UNKNOWN.
If there’s no polynomial model, SIPNAME is set to NOMODEL.
NPOLFILE- (optional) name of npol reference file
NPOLFILE keyword from science file primary header
UNKNOWN if NPOLFILE keyword is missing or invalid but data extensions exist
or NOMODEL
IDCTAB- (optional)IDCTAB keyword from science file primary header or N/A
D2IMFILE- (optional)D2IMFILE keyword from science file primary header or N/A
AUTHOR- (optional) name of person who created the headerlet
DESCRIP- (optional) short description of the headerlet solution
NMATCH- (optional) number of sources used in the new solution fit, if updated from the Archive’s default WCS
CATALOG- (optional) a reference frame used to define the astrometric solution
UPWCSVER- (optional) version of STWCS used to create the WCS of the original image
PYWCSVER- (optional) version of PyWCS used to create the WCS of the original image
These keywords are used to determine whether a headerlet can be applied to a given exposure or not. Some of the keywords provide more information about the solution itself, how it was derived, and by whom.
Working With Headerlets
Headerlets are implemented in a python module headerlet which uses PyWCS for
WCS management and PyFITS for FITS file handling. The functionality includes methods to:
- Create a headerlet (on disk or in memory) from a specific WCS of a science observation.
This can be the Primary or an alternate WCS.
- Apply a WCS from a headerlet to the Primary WCS of a science observation (and
optionally save the original WCS as an alternate WCs or a different headerlet).
Copy a WCS from a headerlet as an alternate WCS.
Attach a headerlet to a science file.
Archive a WCS of a science file as a headerlet attached to the file.
Delete a headerlet attached to a science file.
Print a summary of all headerlets attached to a science file.
An optional GUI interface is available through TEAL and includes functions for writing a headerlet,
applying a headerlet, etc. A full listing of all functions with GUI interface is available
after stwcs.wcsutil is imported.
The headerlet API as of the time of writing this report is documented in Appendix 1: Headerlet API.
Note: For an up-to-date API always consult the current the SSB documentation pages.
Headerlet HDU - A New Type of FITS Extension
The word headerlet has been used sofar in three different ways:
A single WCS representation
The multi-extension FITS file storing a WCS
The extension of a science file containing a headerlet (as a WCS representation)
The last usage of the term headerlet is discussed in this section.
When a headerlet is applied to an image, a copy of the original headerlet file is
appended to the image’s HDU list as a special extension HDU called a HeaderletHDU.
A HeaderletHDU consists of a simple header describing the headerlet, and has as its data
the headerlet file itself, (which may be compressed). A HeaderletHDU has an ‘XTENSION’
value of ‘HDRLET’. Support for this is provided through the implementation of a
NonstandardExtHDU in PyFITS.
When opening a file that contains Headerlet HDU extensions, it will normally look like this in PyFITS:
>>> import pyfits
>>> hdul = pyfits.open('94f05bgq_flt_with_hlet.fits')
>>> hdul.info()
Filename: j94f05bgq_flt_with_hlet.fits
No. Name Type Cards Dimensions Format
0 PRIMARY PrimaryHDU 248 () int16
1 SCI ImageHDU 286 (4096, 2048) float32
2 ERR ImageHDU 76 (4096, 2048) float32
3 DQ ImageHDU 66 (4096, 2048) int16
4 SCI ImageHDU 282 (4096, 2048) float32
5 ERR ImageHDU 74 (4096, 2048) float32
6 DQ ImageHDU 66 (4096, 2048) int16
7 WCSCORR BinTableHDU 56 10R x 23C [40A, I, 1A, D, D, D, D, D, D, D, D, 24A, 24A, D, D, D, D, D, D, D, D, J, 40A]
8 WCSDVARR ImageHDU 15 (65, 33) float32
9 WCSDVARR ImageHDU 15 (65, 33) float32
10 WCSDVARR ImageHDU 15 (65, 33) float32
11 WCSDVARR ImageHDU 15 (65, 33) float32
12 D2IMARR ImageHDU 12 (4096,) float32
13 HDRLET NonstandardExtHDU 13
14 HDRLET NonstandardExtHDU 13
The names of the headerlet extensions are both HDRLET, but its type shows up
as NonstandardExtHDU. Their headers can be read, and while their data can be
read you’d have to know what to do with it (the data is actually
either a tar file or a gzipped tar file containing the
headerlet file). However, if you have stwcs.wcsutil.headerlet imported, PyFITS will
recognize these extensions as Headerlet HDUs:
>>> import stwcs.wcsutil.headerlet
>>> # Note that it's necessary to reopen the file
>>> hdul = pyfits.open('j94f05bgq_flt_with_hlet.fits')
>>> hdul.info()
Filename: j94f05bgq_flt_with_hlet.fits
No. Name Type Cards Dimensions Format
0 PRIMARY PrimaryHDU 248 () int16
1 SCI ImageHDU 286 (4096, 2048) float32
2 ERR ImageHDU 76 (4096, 2048) float32
3 DQ ImageHDU 66 (4096, 2048) int16
4 SCI ImageHDU 282 (4096, 2048) float32
5 ERR ImageHDU 74 (4096, 2048) float32
6 DQ ImageHDU 66 (4096, 2048) int16
7 WCSCORR BinTableHDU 56 10R x 23C [40A, I, 1A, D, D, D, D, D, D, D, D, 24A, 24A, D, D, D, D, D, D, D, D, J, 40A]
8 WCSDVARR ImageHDU 15 (65, 33) float32
9 WCSDVARR ImageHDU 15 (65, 33) float32
10 WCSDVARR ImageHDU 15 (65, 33) float32
11 WCSDVARR ImageHDU 15 (65, 33) float32
12 D2IMARR ImageHDU 12 (4096,) float32
13 HDRLET HeaderletHDU 13
14 HDRLET HeaderletHDU 13
>>> print hdul['HDRLET', 1].header.ascard
XTENSION= 'HDRLET ' / Headerlet extension
BITPIX = 8 / array data type
NAXIS = 1 / number of array dimensions
NAXIS1 = 102400 / Axis length
PCOUNT = 0 / number of parameters
GCOUNT = 1 / number of groups
EXTNAME = 'HDRLET ' / name of the headerlet extension
HDRNAME = 'j94f05bgq_orig' / Headerlet name
DATE = '2011-04-13T12:14:42' / Date FITS file was generated
SIPNAME = 'IDC_qbu1641sj' / SIP distortion model name
NPOLFILE= '/grp/hst/acs/lucas/new-npl/qbu16424j_npl.fits' / Non-polynomial correction
D2IMFILE= '/grp/hst/acs/lucas/new-npl/wfc_ref68col_d2i.fits' / Column correction
COMPRESS= F / Uses gzip compression
HeaderletHDU objects are similar to other HDU objects in PyFITS.
However, they have a special .headerlet attribute that returns
the actual headerlet contained in the HDU data as a Headerlet object:
>>> hdrlet = hdul['HDERLET', 1].headerlet
>>> hdrlet.info()
Filename: (No file associated with this HDUList)
No. Name Type Cards Dimensions Format
0 PRIMARY PrimaryHDU 12 () uint8
1 SIPWCS ImageHDU 111 () uint8
2 SIPWCS ImageHDU 110 () uint8
3 WCSDVARR ImageHDU 15 (65, 33) float32
4 WCSDVARR ImageHDU 15 (65, 33) float32
5 WCSDVARR ImageHDU 15 (65, 33) float32
6 WCSDVARR ImageHDU 15 (65, 33) float32
7 D2IMARR ImageHDU 12 (4096,) float32
This is useful if you want to view the contents of the headerlets attached to a file.
Examples
To create a headerlet from an image, a createHeaderlet() function is provided:
>>> from stwcs.wcsutil import headerlet
>>> hdrlet = headerlet.createHeaderlet('j94f05bgq_flt.fits', 'VERSION1')
>>> type(hdrlet)
<class 'stwcs.wcsutil.headerlet.Headerlet'>
>>> hdrlet.info()
Filename: (No file associated with this HDUList)
No. Name Type Cards Dimensions Format
0 PRIMARY PrimaryHDU 12 ()
1 SIPWCS ImageHDU 111 ()
2 SIPWCS ImageHDU 110 ()
3 WCSDVARR ImageHDU 15 (65, 33) float32
4 WCSDVARR ImageHDU 15 (65, 33) float32
5 WCSDVARR ImageHDU 15 (65, 33) float32
6 WCSDVARR ImageHDU 15 (65, 33) float32
7 D2IMARR ImageHDU 12 (4096,) float32
As you can see, the Headerlet object is similar to a normal PyFITS HDUList object. createHeaderlet() can be given either the path
to a file, or an already open HDUList as its first argument.
What do you do with a Headerlet object? Its main purpose is to apply its WCS solution to another file. This can be done using the
Headerlet.apply() method:
>>> hdrlet.apply('some_other_image.fits')
Or you can use the applyHeaderlet() convenience function. It takes an existing headerlet file path or object as its first argument;
the rest of its arguments are the same as Headerlet.apply(). As with createHeaderlet() both of these can take a file path or opened
HDUList objects as arguments.
When a headerlet is applied to an image, an additional headerlet containing that image’s original WCS solution is automatically created,
and is appended to the file’s HDU list as a Headerlet HDU. However, this behavior can be disabled by setting the createheaderlet keyword
argument to False in either Headerlet.apply() or applyHeaderlet().
Appendix 1: Headerlet API
apply_headerlet_as_alternate
def apply_headerlet_as_alternate(filename, hdrlet, attach=True, wcskey=None,
wcsname=None, logging=False, logmode='w'):
"""
Apply headerlet to a science observation as an alternate WCS
Parameters
----------
filename: string
File name of science observation whose WCS solution will be updated
hdrlet: string
Headerlet file
attach: boolean
flag indicating if the headerlet should be attached as a
HeaderletHDU to fobj. If True checks that HDRNAME is unique
in the fobj and stops if not.
wcskey: string
Key value (A-Z, except O) for this alternate WCS
If None, the next available key will be used
wcsname: string
Name to be assigned to this alternate WCS
WCSNAME is a required keyword in a Headerlet but this allows the
user to change it as desired.
logging: boolean
enable file logging
logmode: 'a' or 'w'
"""
apply_headerlet_as_primary
def apply_headerlet_as_primary(filename, hdrlet, attach=True, archive=True,
force=False, logging=False, logmode='a'):
"""
Apply headerlet 'hdrfile' to a science observation 'destfile' as the primary WCS
Parameters
----------
filename: string
File name of science observation whose WCS solution will be updated
hdrlet: string
Headerlet file
attach: boolean
True (default): append headerlet to FITS file as a new extension.
archive: boolean
True (default): before updating, create a headerlet with the
WCS old solution.
force: boolean
If True, this will cause the headerlet to replace the current PRIMARY
WCS even if it has a different distortion model. [Default: False]
logging: boolean
enable file logging
logmode: 'w' or 'a'
log file open mode
"""
archive_as_headerlet
def archive_as_headerlet(filename, hdrname, sciext='SCI',
wcsname=None, wcskey=None, destim=None,
sipname=None, npolfile=None, d2imfile=None,
author=None, descrip=None, history=None,
nmatch=None, catalog=None,
logging=False, logmode='w'):
"""
Save a WCS as a headerlet extension and write it out to a file.
This function will create a headerlet, attach it as an extension to the
science image (if it has not already been archived) then, optionally,
write out the headerlet to a separate headerlet file.
Either wcsname or wcskey must be provided, if both are given, they must match a valid WCS
Updates wcscorr if necessary.
Parameters
----------
filename: string or HDUList
Either a filename or PyFITS HDUList object for the input science file
An input filename (str) will be expanded as necessary to interpret
any environmental variables included in the filename.
hdrname: string
Unique name for this headerlet, stored as HDRNAME keyword
sciext: string
name (EXTNAME) of extension that contains WCS to be saved
wcsname: string
name of WCS to be archived, if " ": stop
wcskey: one of A...Z or " " or "PRIMARY"
if " " or "PRIMARY" - archive the primary WCS
destim: string
DESTIM keyword
if NOne, use ROOTNAME or science file name
sipname: string or None (default)
Name of unique file where the polynomial distortion coefficients were
read from. If None, the behavior is:
The code looks for a keyword 'SIPNAME' in the science header
If not found, for HST it defaults to 'IDCTAB'
If there is no SIP model the value is 'NOMODEL'
If there is a SIP model but no SIPNAME, it is set to 'UNKNOWN'
npolfile: string or None (default)
Name of a unique file where the non-polynomial distortion was stored.
If None:
The code looks for 'NPOLFILE' in science header.
If 'NPOLFILE' was not found and there is no npol model, it is set to 'NOMODEL'
If npol model exists, it is set to 'UNKNOWN'
d2imfile: string
Name of a unique file where the detector to image correction was
stored. If None:
The code looks for 'D2IMFILE' in the science header.
If 'D2IMFILE' is not found and there is no d2im correction,
it is set to 'NOMODEL'
If d2im correction exists, but 'D2IMFILE' is missing from science
header, it is set to 'UNKNOWN'
author: string
Name of user who created the headerlet, added as 'AUTHOR' keyword
to headerlet PRIMARY header
descrip: string
Short description of the solution provided by the headerlet
This description will be added as the single 'DESCRIP' keyword
to the headerlet PRIMARY header
history: filename, string or list of strings
Long (possibly multi-line) description of the solution provided
by the headerlet. These comments will be added as 'HISTORY' cards
to the headerlet PRIMARY header
If filename is specified, it will format and attach all text from
that file as the history.
logging: boolean
enable file folling
logmode: 'w' or 'a'
log file open mode
"""
attach_headerlet
def attach_headerlet(filename, hdrlet, logging=False, logmode='a'):
"""
Attach Headerlet as an HeaderletHDU to a science file
Parameters
----------
filename: string, HDUList
science file to which the headerlet should be applied
hdrlet: string or Headerlet object
string representing a headerlet file
logging: boolean
enable file logging
logmode: 'a' or 'w'
"""
create_headerlet
def create_headerlet(filename, sciext='SCI', hdrname=None, destim=None,
wcskey=" ", wcsname=None,
sipname=None, npolfile=None, d2imfile=None,
author=None, descrip=None, history=None,
nmatch=None, catalog=None,
logging=False, logmode='w'):
"""
Create a headerlet from a WCS in a science file
If both wcskey and wcsname are given they should match, if not
raise an Exception
Parameters
----------
filename: string or HDUList
Either a filename or PyFITS HDUList object for the input science file
An input filename (str) will be expanded as necessary to interpret
any environmental variables included in the filename.
sciext: string or python list (default: 'SCI')
Extension in which the science data with the linear WCS is.
The headerlet will be created from these extensions.
If string - a valid EXTNAME is expected
If int - specifies an extension with a valid WCS, such as 0 for a
simple FITS file
If list - a list of FITS extension numbers or strings representing
extension tuples, e.g. ('SCI, 1') is expected.
hdrname: string
value of HDRNAME keyword
Takes the value from the HDRNAME<wcskey> keyword, if not available from WCSNAME<wcskey>
It stops if neither is found in the science file and a value is not provided
destim: string or None
name of file this headerlet can be applied to
if None, use ROOTNAME keyword
wcskey: char (A...Z) or " " or "PRIMARY" or None
a char representing an alternate WCS to be used for the headerlet
if " ", use the primary (default)
if None use wcsname
wcsname: string or None
if wcskey is None use wcsname specified here to choose an alternate WCS for the headerlet
sipname: string or None (default)
Name of unique file where the polynomial distortion coefficients were
read from. If None, the behavior is:
The code looks for a keyword 'SIPNAME' in the science header
If not found, for HST it defaults to 'IDCTAB'
If there is no SIP model the value is 'NOMODEL'
If there is a SIP model but no SIPNAME, it is set to 'UNKNOWN'
npolfile: string or None (default)
Name of a unique file where the non-polynomial distortion was stored.
If None:
The code looks for 'NPOLFILE' in science header.
If 'NPOLFILE' was not found and there is no npol model, it is set to 'NOMODEL'
If npol model exists, it is set to 'UNKNOWN'
d2imfile: string
Name of a unique file where the detector to image correction was
stored. If None:
The code looks for 'D2IMFILE' in the science header.
If 'D2IMFILE' is not found and there is no d2im correction,
it is set to 'NOMODEL'
If d2im correction exists, but 'D2IMFILE' is missing from science
header, it is set to 'UNKNOWN'
author: string
Name of user who created the headerlet, added as 'AUTHOR' keyword
to headerlet PRIMARY header
descrip: string
Short description of the solution provided by the headerlet
This description will be added as the single 'DESCRIP' keyword
to the headerlet PRIMARY header
history: filename, string or list of strings
Long (possibly multi-line) description of the solution provided
by the headerlet. These comments will be added as 'HISTORY' cards
to the headerlet PRIMARY header
If filename is specified, it will format and attach all text from
that file as the history.
nmatch: int (optional)
Number of sources used in the new solution fit
catalog: string (optional)
Astrometric catalog used for headerlet solution
logging: boolean
enable file logging
logmode: 'w' or 'a'
log file open mode
Returns
-------
Headerlet object
"""
delete_headerlet
def delete_headerlet(filename, hdrname=None, hdrext=None, distname=None,
logging=False, logmode='w'):
"""
Deletes HeaderletHDU(s) from a science file
Notes
-----
One of hdrname, hdrext or distname should be given.
If hdrname is given - delete a HeaderletHDU with a name HDRNAME from fobj.
If hdrext is given - delete HeaderletHDU in extension.
If distname is given - deletes all HeaderletHDUs with a specific distortion model from fobj.
Updates wcscorr
Parameters
----------
filename: string or HDUList
Either a filename or PyFITS HDUList object for the input science file
An input filename (str) will be expanded as necessary to interpret
any environmental variables included in the filename.
hdrname: string or None
HeaderletHDU primary header keyword HDRNAME
hdrext: int, tuple or None
HeaderletHDU FITS extension number
tuple has the form ('HDRLET', 1)
distname: string or None
distortion model as specified in the DISTNAME keyword
logging: boolean
enable file logging
logmode: 'a' or 'w'
"""
extract_headerlet
def extract_headerlet(filename, output, extnum=None, hdrname=None,
clobber=False, logging=True):
"""
Finds a headerlet extension in a science file and writes it out as
a headerlet FITS file.
If both hdrname and extnum are given they should match, if not
raise an Exception
Parameters
----------
filename: string or HDUList or Python list
This specifies the name(s) of science file(s) from which headerlets
will be extracted.
String input formats supported include use of wild-cards, IRAF-style
'@'-files (given as '@<filename>') and comma-separated list of names.
An input filename (str) will be expanded as necessary to interpret
any environmental variables included in the filename.
If a list of filenames has been specified, it will extract a
headerlet from the same extnum from all filenames.
output: string
Filename or just rootname of output headerlet FITS file
If string does not contain '.fits', it will create a filename with
'_hlet.fits' suffix
extnum: int
Extension number which contains the headerlet to be written out
hdrname: string
Unique name for headerlet, stored as the HDRNAME keyword
It stops if a value is not provided and no extnum has been specified
clobber: bool
If output file already exists, this parameter specifies whether or not
to overwrite that file [Default: False]
logging: boolean
enable logging to a file
"""
print_summary
def print_summary(summary_cols, summary_dict, pad=2, maxwidth=None, idcol=None, output=None, clobber=True, quiet=False ): """ Print out summary dictionary to STDOUT, and possibly an output file """
restore_all_with_distname
def restore_all_with_distname(filename, distname, primary, archive=True,
sciext='SCI', logging=False, logmode='w'):
"""
Restores all HeaderletHDUs with a given distortion model as alternate WCSs and a primary
Parameters
--------------
filename: string or HDUList
Either a filename or PyFITS HDUList object for the input science file
An input filename (str) will be expanded as necessary to interpret
any environmental variables included in the filename.
distname: string
distortion model as represented by a DISTNAME keyword
primary: int or string or None
HeaderletHDU to be restored as primary
if int - a fits extension
if string - HDRNAME
if None - use first HeaderletHDU
archive: boolean (default True)
flag indicating if HeaderletHDUs should be created from the
primary and alternate WCSs in fname before restoring all matching
headerlet extensions
logging: boolean
enable file logging
logmode: 'a' or 'w'
"""
restore_from_headerlet
def restore_from_headerlet(filename, hdrname=None, hdrext=None, archive=True,
force=False, logging=False, logmode='w'):
"""
Restores a headerlet as a primary WCS
Parameters
----------
filename: string or HDUList
Either a filename or PyFITS HDUList object for the input science file
An input filename (str) will be expanded as necessary to interpret
any environmental variables included in the filename.
hdrname: string
HDRNAME keyword of HeaderletHDU
hdrext: int or tuple
Headerlet extension number of tuple ('HDRLET',2)
archive: boolean (default: True)
When the distortion model in the headerlet is the same as the distortion model of
the science file, this flag indicates if the primary WCS should be saved as an alternate
nd a headerlet extension.
When the distortion models do not match this flag indicates if the current primary and
alternate WCSs should be archived as headerlet extensions and alternate WCS.
force: boolean (default:False)
When the distortion models of the headerlet and the primary do not match, and archive
is False, this flag forces an update of the primary.
logging: boolean
enable file logging
logmode: 'a' or 'w'
"""
write_headerlet
def write_headerlet(filename, hdrname, output=None, sciext='SCI',
wcsname=None, wcskey=None, destim=None,
sipname=None, npolfile=None, d2imfile=None,
author=None, descrip=None, history=None,
nmatch=None, catalog=None,
attach=True, clobber=False, logging=False):
"""
Save a WCS as a headerlet FITS file.
This function will create a headerlet, write out the headerlet to a
separate headerlet file, then, optionally, attach it as an extension
to the science image (if it has not already been archived)
Either wcsname or wcskey must be provided; if both are given, they must
match a valid WCS.
Updates wcscorr if necessary.
Parameters
----------
filename: string or HDUList or Python list
This specifies the name(s) of science file(s) from which headerlets
will be created and written out.
String input formats supported include use of wild-cards, IRAF-style
'@'-files (given as '@<filename>') and comma-separated list of names.
An input filename (str) will be expanded as necessary to interpret
any environmental variables included in the filename.
hdrname: string
Unique name for this headerlet, stored as HDRNAME keyword
output: string or None
Filename or just rootname of output headerlet FITS file
If string does not contain '.fits', it will create a filename
starting with the science filename and ending with '_hlet.fits'.
If None, a default filename based on the input filename will be
generated for the headerlet FITS filename
sciext: string
name (EXTNAME) of extension that contains WCS to be saved
wcsname: string
name of WCS to be archived, if " ": stop
wcskey: one of A...Z or " " or "PRIMARY"
if " " or "PRIMARY" - archive the primary WCS
destim: string
DESTIM keyword
if NOne, use ROOTNAME or science file name
sipname: string or None (default)
Name of unique file where the polynomial distortion coefficients were
read from. If None, the behavior is:
The code looks for a keyword 'SIPNAME' in the science header
If not found, for HST it defaults to 'IDCTAB'
If there is no SIP model the value is 'NOMODEL'
If there is a SIP model but no SIPNAME, it is set to 'UNKNOWN'
npolfile: string or None (default)
Name of a unique file where the non-polynomial distortion was stored.
If None:
The code looks for 'NPOLFILE' in science header.
If 'NPOLFILE' was not found and there is no npol model, it is set to 'NOMODEL'
If npol model exists, it is set to 'UNKNOWN'
d2imfile: string
Name of a unique file where the detector to image correction was
stored. If None:
The code looks for 'D2IMFILE' in the science header.
If 'D2IMFILE' is not found and there is no d2im correction,
it is set to 'NOMODEL'
If d2im correction exists, but 'D2IMFILE' is missing from science
header, it is set to 'UNKNOWN'
author: string
Name of user who created the headerlet, added as 'AUTHOR' keyword
to headerlet PRIMARY header
descrip: string
Short description of the solution provided by the headerlet
This description will be added as the single 'DESCRIP' keyword
to the headerlet PRIMARY header
history: filename, string or list of strings
Long (possibly multi-line) description of the solution provided
by the headerlet. These comments will be added as 'HISTORY' cards
to the headerlet PRIMARY header
If filename is specified, it will format and attach all text from
that file as the history.
attach: bool
Specify whether or not to attach this headerlet as a new extension
It will verify that no other headerlet extension has been created with
the same 'hdrname' value.
clobber: bool
If output file already exists, this parameter specifies whether or not
to overwrite that file [Default: False]
logging: boolean
enable file logging
"""
Hack, et al, STScI 2012-01 TSR, http://stsdas.stsci.edu/tsr
(draft FITS WCS Distortion paper) Calabretta M. R., Valdes F. G., Greisen E. W., and Allen S. L., 2004, “Representations of distortions in FITS world coordinate systems”,[cited 2012 Sept 18], Available from: http://www.atnf.csiro.au/people/mcalabre/WCS/dcs_20040422.pdf